Introduction
Corynebacterium pseudodiphtheriticum, a member of genus Corynebacterium, is a common upper respiratory tract commensal. However, this bacterium is increasingly being reported as a cause of various infections ranging from upper and lower respiratory tract infections, endocarditis, cutaneous and ocular infections [1,2]. Most of these patients are immunocompromised or have other comorbidities [1,3]. C. pseudodiphtheriticum can cause exudative pharyngitis which sometimes may mimic diphtheria [4,5]. This can pose a challenge for the treating physicians, especially in the setting of a diphtheria epidemic.
Materials and Methods
A retrospective descriptive study was carried out at the Department of Microbiology, Government Medical College, Manjeri, Kerala, India from January 2017 and July 2017. A waiver of informed consent was obtained from Institutional Ethics Committee (IEC/GMCM/09/17). Patient anonymity was strictly maintained throughout the study. Patient details were collected from laboratory and medical records.
Throat swabs collected from clinically suspected cases of diphtheria were included in the study. swabs sent for other reasons like follicular tonsillitis were excluded. Cases were determined as ‘suspected’ according to the interim guidelines released by the Government of Kerala [6].
Culture and Identification Procedures
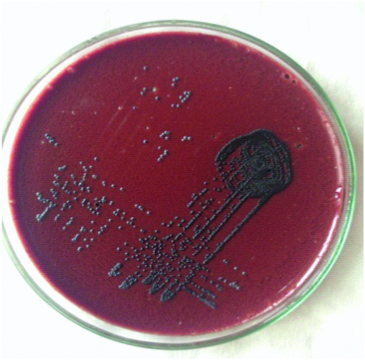
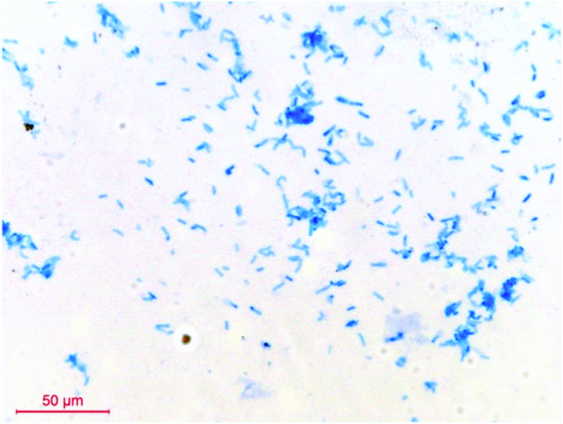
Throat swabs were collected from each patient. Gram’s smears were examined for predominant presence of Gram positive bacilli. Swabs were inoculated onto Blood Agar (BA) and Potassium Tellurite Blood Agar (PTBA) and incubated aerobically at 37oC for 48 hours. The BA plates were checked for the presence of white, opaque, non-haemolytic colonies and PTBA plates for black colonies [Table/Fig-1,2]. Only those specimens which yielded pure growth of black colonies on PTBA were included in the study. Gram’s and methylene blue smears were prepared from the growth [Table/Fig-3,4]. Colonies of Gram positive bacilli with cuneiform arrangement and metachromatic granules were reported provisionally as ‘Organisms morphologically resembling C. diphtheriae grown in culture’.
Opaque, grey, smooth, convex non haemolytic colonies on blood agar after 48 hours incubation at 38°C.

Slightly dry, convex, grey/black colonies on Potassium tellurite blood agar after 48 hours incubation at 38°C.
Gram’s stain (100X) C. pseudodophtheriticum-Gram positive bacilli in cuneiform arrangement.

Methylene blue stain (100X) C. pseudodophtheriticum-metachromatic granules and cuneiform arrangement.
All isolates morphologically resembling Corynebacterium diphtheriae were sent to State Public Health Laboratory (SPHL) Thiruvananthapuram for further identification and ‘tox’ gene study. At SPHL, identification was done by standard biochemical tests [7]. Polymerase Chain Reaction (PCR) for the presence of ‘tox’ gene was done by amplification of specific sequences using primers described previously [8]. Elek’s test was performed to confirm the expression of ‘tox’ genes using modified Elek’s test protocol [9]. We received the final report from SPHL within 7-10 days.
Antibiotic Sensitivity Testing (ABST) was done on Mueller Hinton agar supplemented with 5% blood by Kirby Bauer method. As final identification of the isolates took about 7-10 days, we proceeded with ABST on ‘Organisms morphologically resembling C. diphtheriae’ considering the gravity of the disease. Inoculum was prepared according to Clinical and Laboratory Standards Institute (CLSI) guidelines [10]. Penicillin (10 U), Ampicillin (10 μg), Erythromycin (15 μg) and Azithromycin (15 μg) discs (Hi Media) were used for testing. CLSI do not provide zone diameter breakpoints for disc diffusion method of antibiotic sensitivity testing for Corynebacterium spp. CLSI determined breakpoints for staphylococci were used for interpretation [11,12].
After collecting throat swabs, patients were given Erythromycin (40 mg/Kg/day in four divided doses) for 14 days from the outpatient clinics. Immediately after receiving the presumptive report of a morphological diagnosis, the patients were called back by the primary physicians and were referred to Government Medical College, Kozhikode for further management as the hospital did not have the facility to administer Anti-diphtheria antiserum in required cases. Erythromycin was discontinued and patients were treated with Crystalline Penicillin (200000 IU/Kg in four divided doses) for 14 days.
Few isolates resembling C.diphtheriae obtained between May-July 2017 were resistant to macrolides but sensitive to Penicillin and Ampicillin. These isolates were sent to Aster MIMS Microbiology laboratory for identification and ‘tox’ gene detection by PCR. Species identification was done by VITEC-2 Compact system (bioMérieux) and ‘tox’ gene detection by PCR [8]. Antibiotic sensitivity tests were done with E-strips (bioMérieux) for Penicillin (0.016-32 μg/mL), Ampicillin (0.016-32 μg/mL), Erythromycin (0.016-32 μg/mL), Azithromycin (0.016-32 μg/mL) and Clindamycin (0.016-32 μg/mL). The CLSI determined breakpoints were used for interpreting MIC of Penicillin, Erythromycin and Clindamycin [10]. For Ampicillin and Azithromycin, CLSI determined breakpoints for staphylococci have been used [11].
Results
A total of 341 throat swabs were processed between January 2017 and July 2017 of which 152 were from males and 189 from females.
Of the 341 suspected cases, a provisional report of ‘Organisms morphologically resembling C. diphtheriae have grown in culture’ was issued for 33 (9.6%) patients. A total of 20 isolates among these were confirmed as C. diphtheriae from SPHL of which 17 were toxigenic strains. Three isolates were identified as C. minutissimum, one C. jeikeium and one Arcanobacterium.
A total of 8 out of 33 isolates were identified as C. pseudodiphtheriticum. All tested negative for the presence of ‘tox’ gene [Table/Fig-5]. All eight patients were children below 10 years of age and had presented to the outpatient department with features of fever and exudative pharyngitis. Apart from the current illness all eight children were healthy and there was no history suggestive of a compromised immune system.
Lanes1-8 test strains C. pseudodiphtheriticum. Negative for ‘tox’ gene. 150 bp denotes ‘tox’ gene.
PC: Positive control, NC: Negative control

All patients recovered from the illness. Details like age, sex, primary immunisation status, presenting complaints and presence or absence of signs of toxicity of eight patients from whom C. pseudodiphtheriticum were isolated are given in [Table/Fig-6].
Clinical details of 8 patients including immunisation status.
| S.No | Age/Sex | Immunisation status | Fever | Sore throat | Pseudomembrane | Clinical signs of toxicity | ADS*given |
|---|
| 1 | 10/M | Fully immunised | + | + | + R tonsil | _ | _ |
| 2 | 10/M | Partially immunised (till 1.5 years) | + | + | + | _ | +§ |
| 3 | 8/M | No records available | + | + | + R tonsil | _ | +§ |
| 4 | 10/F | Partially immunised (till 1.5 years) | + | + | + | _ | _ |
| 5 | 10/F | Completed (till 5 years) | + | + | + | _ | _ |
| 6 | 7/F | Fully immunised | + | + | + | _ | _ |
| 7 | 9/M | Fully immunised | + | + | + | _ | _ |
| 8 | 10/F | Partially immunised (till 1.5 years) | + | + | + R tonsil | _ | _ |
M: Male; F: Female
*Anti diphtheria antiserum
§Patients who received ADS
Two patients received ADS, but discontinued due to hypersensitivityreaction
Discussion
The Northern districts of Kerala, especially Malappuram and Kozhikode witnessed a resurgence of diphtheria which started in June 2015. By 2016 the numbers of diphtheria cases rose to epidemic proportions [13]. Government Medical College, Manjeri serves a large number of patients belonging to Malappuram district. The index case during the 2015 outbreak was reported from this district. All suspected cases of diphtheria from this area were being referred to Government Medical College, Kozhikode till June 2016 after which laboratory started basic culture of throat swabs for diphtheria.
Diagnosis of diphtheria was made based on clinical features including the presence of a pseudo-membrane [6]. Swabs collected from suspected cases of diphtheria were sent to microbiology laboratory for culture. Although membranous pharyngitis can be caused by several organisms including bacteria, viruses and Candida, diphtheria remains the first differential diagnosis in the setting of an epidemic [4].
During the study period, 33 isolates were reported presumptively as C. diphtheriae of which 8 (24%) were identified as C pseudodiphtheriticum. All were from children below 10 years of age. Among them four were males and four were females. Pathogenic potential of C. pseudodiphtheriticum is well known but is usually associated with an immunocompromised state or other underlying illness [14-16]. All the eight patients included in this study presented with signs and symptoms of severe upper respiratory tract illness. There was no history of previous chronic illnesses, invasive procedures or transplants. Metachromatic granules are absent or are minimally seen in C. pseudodiphtheriticum [17]. Among the eight isolates in present series, five had minimal metachromatic granules. With prolonged incubation, granules were more prominent. In the eight cases that we have reported, C. pseudodiphtheriticum was grown in pure culture in PTBA and was the predominant growth in BA. This confirms the association of the isolate with exudative pharyngitis. All isolates were negative for tox gene; hence, the pathogenicity cannot be attributed to toxins. Though, a common commensal of upper respiratory tract, C. pseudodiphtheriticum has the potential to breach epithelial cell barrier, invade deeper tissue and elicit pro-inflammatory responses which could explain the signs and symptoms [4,18].
Although, the beginning of the current outbreak was in 2015, C. pseudodiphtheriticum was isolated for the first time in May 2017 with clustering of cases till July. From August 2017-till date we have not isolated C.pseudodiphtheriticum even though we continue to isolate C. diphtheriae. C. pseudodiphtheriticum shows variable susceptibility to macrolides and lincosamides, whereas susceptibility to beta lactams has been near universal [1,2,19]. In the present study all isolates were sensitive to beta lactams; however resistance to macrolides and clindamycin was 100%. Resistance to macrolides is reported in other Corynebacterium spp. like C. amycolatum, C. urealyticum, C. afermentans and C. auris. C. jeikeium shows inducible resistance to macrolides [3]. C. minutissimum and C. jeikeium isolated from the present patients were sensitive to all antibiotics tested. Resistance to beta lactam or macrolides has not been reported in C. diphtheriae isolated from North Kerala during the present outbreak since 2015. In this scenario, resistance to macrolides should raise a suspicion that the isolate may not be C. diphtheriae.
For the eight patients with a presumptive diagnosis of diphtheria, prompt treatment had to be initiated according to the protocol without waiting for lab confirmation which took about 7-10 days. Patients had to be shifted to a hospital in a different town anticipating treatment with ADS. ADS administered for two out of eight patients had to be discontinued due to hypersensitivity reactions. In addition to the considerable inconvenience incurred by patients and family, administrative machinery had to be mobilised for tracing contacts to institute prophylactic treatment which was not required, had a final confirmation of the isolate been possible without time delay.
Microbiology laboratories in public sector centres catering to this area need to be equipped with facilities for rapid confirmation of identity of isolates so that unnecessary delay in diagnosis, unwanted hospital admissions and treatments can be avoided.
Limitation
We were unable to confirm the identity of the probable Corynebacterium isolates before proceeding to antibiotic sensitivity tests due to resource constraints. There was clustering of isolates during the months of May, June and July. We did not investigate whether these patients came from the same locality. Epidemiological typing was not done to establish clonal origin of the isolates.
Conclusion
C. pseudodiphtheriticum presenting as exudative pharyngitis can mimic mild diphtheria clinically and pose a diagnostic challenge in the setting of a diphtheria epidemic. Macrolide resistance provides a clue that the isolate may not be C. diphtheriae.
M: Male; F: Female
*Anti diphtheria antiserum
§Patients who received ADSTwo patients received ADS, but discontinued due to hypersensitivityreaction
[1]. Van Roeden SE, Thijsen SF, Sankatsing SU, Limonard SJ, Clinical relevance of Corynebacterium pseudodiphtheriticum in lower respiratory tract specimensInfect Dis (Lond) 2015 47(12):862-68.10.3109/23744235.2015.107096226211497 [Google Scholar] [CrossRef] [PubMed]
[2]. Ahmed K, Kawakami K, Watanabe K, Mitsushima H, Nagatake T, Matsumoto K, Corynebacterium pseudodiphtheriticum: a respiratory tract pathogenClin Infect Dis 1995 20(1):41-46.10.1093/clinids/20.1.417727668 [Google Scholar] [CrossRef] [PubMed]
[3]. Bennett JE, Dolin R, Blaser MJ, Chapter 207, Other Coryneform Bacteria and Rhodococci. In: Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases 2015 PhiladelphiaPA: Elsevier/Saunders:2373-2382. [Google Scholar]
[4]. Indumathi VA, Shikha R, Suryaprakash DR, Diphtheria-like illness in a fully immunised child caused by Corynebacterium pseudodiphtheriticumIndian J Med Microbiol 2014 32(4):443-45.10.4103/0255-0857.14225025297035 [Google Scholar] [CrossRef] [PubMed]
[5]. Izurieta HS, Strebel PM, Youngblood T, Hollis DG, Popovic T, Exudative pharyngitis possibly due to Corynebacterium pseudodiphtheriticum, a new challenge in the differential diagnosis of diphtheriaEmerg Infect Dis 1997 3(1):65-68.10.3201/eid0301.9701099126447 [Google Scholar] [CrossRef] [PubMed]
[6]. Diphtheria -Interim Guidelines Kerala Public Health Division. (accessed 26 November 2017). http://dhs.kerala.gov.in/docs/transfer/addlph/25072016dh2.pdf [Google Scholar]
[7]. Procop GW, Church DL, Hall GS, Capter 14, Aerobic and Facultative Gram-Positive Bacilli. In: Koneman’s Color Atlas and Textbook of Diagnostic Microbiology 2016 PhiladelphiaWolters Kluwer Health:843-959. [Google Scholar]
[8]. Nakao H, Popovic T, Development of a direct PCR assay for detection of the diphtheria toxin geneJ Clin Microbiol 1997 35(7):1651-55. [Google Scholar]
[9]. Engler KH, Glushkevich T, Mazurova IK, George RC, Efstratiou A, A modified Elek test for detection of toxigenic corynebacteria in the diagnostic laboratoryJ Clin Microbiol 1997 35(7):495-98. [Google Scholar]
[10]. Jorgensen JH, Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria; approved guideline M45-A2Second Wayne: Clinical and laboratory standards Institute 2010 [Google Scholar]
[11]. Winn WC, Allen C, Janda W, Chapter 14, Aerobic and Facultative Gram Positive Bacilli. In: Koneman’s color atlas and textbook of diagnostic microbiology 2006 Philadelphiaippincott Williams & Wilkins:765-857. [Google Scholar]
[12]. Funke G, Von Graevenit A, Clarridge J, Bernard KA, Clinical microbiology of coryneform bacteriaClin Microbiol Rev 1997 10(10):125-59.10.1128/CMR.10.1.1258993861 [Google Scholar] [CrossRef] [PubMed]
[13]. Sangal L, Joshi S, Anandan S, Balaji V, J Johnson, Satapathy A, Resurgence of diphtheria in North Kerala, India, 2016: laboratory supported case-based surveillance outcomesFront Public Health 2017 5:21810.3389/fpubh.2017.0021828913330 [Google Scholar] [CrossRef] [PubMed]
[14]. Nishiyama A, Ishida T, Ito A, Arita M, Bronchopneumonia caused by Corynebacterium pseudodiphtheriticumIntern Med 2013 52(16):184710.2169/internalmedicine.52.956023955623 [Google Scholar] [CrossRef] [PubMed]
[15]. Camello TC, Souza MC, Martins CA, Damasco PV, Marques EA, Pimenta FP, Corynebacterium pseudodiphtheriticum isolated from relevant clinical sites of infection: a human pathogen overlooked in emerging countriesLett ApplMicrobiol 2009 48(4):458-64.10.1111/j.1472-765X.2009.02553.x19228291 [Google Scholar] [CrossRef] [PubMed]
[16]. Morris A, Guild I, Endocarditis due to Corynebacterium pseudodiphtheriticum: five case reports, review, and antibiotic susceptibilities of nine strainsRev Infect Dis 1991 13(5):887-92.10.1093/clinids/13.5.8871962103 [Google Scholar] [CrossRef] [PubMed]
[17]. Goodfellow M, Whitman WB, Bergey WB, Family 1. Corynebacteriaceae. In: Bergey’s Manual of Systematic Bacteriology:The Actinobacteria 2012 New YorkSpringer:244-300.10.1007/978-0-387-68233-4 [Google Scholar] [CrossRef]
[18]. Roy S, Marla S, Praneetha DC, Recognition of corynebacterium pseudodiphtheriticum by toll-like receptors and up-regulation of antimicrobial peptides in human corneal epithelial cellsVirulence 2015 6(7):716-21.10.1080/21505594.2015.106606326125127 [Google Scholar] [CrossRef] [PubMed]
[19]. Manzella JP, Kellogg JA, Parsey KS, Corynebacterium pseudodiphtheriticum: a respiratory tract pathogen in adultsClin Infect Dis 1995 20(1):37-40.10.1093/clinids/20.1.377727667 [Google Scholar] [CrossRef] [PubMed]