Phenotypic and Genotypic Methods for Detection of Extended Spectrum β Lactamase Producing Escherichia coli and Klebsiella pneumoniae Isolated from Ventilator Associated Pneumonia
Veena Krishnamurthy1, Vijaykumar G.S.2, Sudeepa Kumar M.3, Prashanth H.V.4, Prakash R.5, Nagaraj E.R.6
1 Assistant Professor, Department of Microbiology, Sri Siddhartha Medical College, Agalakote, Tumkur-572107, Karnataka, India.
2 Professor and HOD, Department of Microbiology, J.S.S. Medical College, Mysore, Karnataka, India.
3 Assistant Professor, Department of Microbiology, Sri Siddhartha Medical College, Agalakote, Tumkur-572107, Karnataka, India.
4 Professor, Department of Microbiology, Sri Siddhartha Medical College, Agalakote, Tumkur-572107, Karnataka, India.
5 Assistant Professor, Department of Microbiology, Sri Siddhartha Medical College, Agalakote, Tumkur-572107, Karnataka, India.
6 Professor and HOD, Department of Microbiology, Sri Siddhartha Medical College, Agalakote, Tumkur-572107, Karnataka, India.
NAME, ADDRESS, E-MAIL ID OF THE CORRESPONDING AUTHOR: Dr. Veena Krishnamurthy, Assistant Professor, Department of Microbiology, Sri Siddhartha Medical College, Agalakote, Tumkur-572107, Karnataka, India.
Phone: 9886432857,
E-mail: veena_kumara@rediffmail.com
Background: Ventilator Associated Pneumonia (VAP) is one of the common nosocomial infections associated with high morbidity due to multidrug resistant pathogens. Rapid spread of resistance to broad spectrum beta-lactams in pathogenic strains causes antibiotics ineffectiveness and increased severity of illness. The CTX-M is the most dominant Extended Spectrum β Lactamase (ESBL) among Enterobacteriaceae in many regions of the world. The aim of the study was to identify the occurrence of ESBL and detect the genes responsible for ESBL production by conventional Polymerase Chain Reaction (PCR) method.
Methods: This prospective study included patients, clinically diagnosed as VAP. Endotracheal aspirates (EA) were collected and cultured by quantitative method. The bacterial isolates were identified as per standard methods. Isolates resistant to 3rd generation cephalosporins were screened for ESBL production by disk approximation method and combination disc diffusion method. Isolates confirmed as ESBL producers were subjected to genotyping by conventional PCR.
StatisticalAnalysis: Statistical analysis was done by using MS Excel sheet. Descriptive statistics like percentage was done in the study.
Results: Among the isolates from 428 patients who developed VAP, 144 isolates belonged to the Enterobacteriaceae family (Klebsiella pneumoniae 87 and Escherichia coli 57). A total of 66 isolates (28 Klebsiella pneumoniae and 38 Escherichia coli) were confirmed as ESBL producer by disc approximation method and 63 isolates by double disc combination method. In the present study by conventional PCR bla CTX-M was the common gene in 48.5% strains followed by 22.22% bla SHV and 14.81% bla TEM.
Conclusion: The genotypic methods using specific PCR amplification of resistance genes seems to have 100% specificity and sensitivity in detection of ESBL when compared to phenotypic methods which lacks the constant sensitivity.
blaTEM gene, blaSHV gene, blaCTX-M gene, Extended Spectrum Beta Lactamase, Polymerase Chain Reaction, Ventilator Associated Pneumonia
Introduction
Ventilator Associated Pneumonia (VAP) is one of the common nosocomial infections among patients admitted to the intensive care unit (ICU) [1] associated with high morbidity and mortality [2]. Risk of mortality and morbidity in VAP is increased due to wrong or delayed initial antibiotic therapy especially when VAP is caused by multidrug resistant pathogens (MDR) [3]. One of these resistant pathogens are the extended spectrum β- lactamase (ESBL) producing Escherichia coli and Klebsiella pneumoniae which have spread worldwide since their discovery in 1983 and pose a serious threat in healthcare associated infections [4]. ESBL producing strains often exhibit multidrug resistance, including resistance to aminoglycosides and fluoroquinolones, limiting the therapeutic options. The typical character of ESBL is their ability to hydrolyse oxyimino-cephalosporins and aztreonam while being inhibited by β-lactamase inhibitors [5]. ESBLs are often plasmid mediated and most are mutants of the classic TEM or SHV β-lactamase enzymes (class A), with one or more amino acid substitutions around the active site [6]. In recent years non TEM and non SHV plasmid mediated ESBL have been reported mainly of the CTX-M type. The CTX-M family first described in the early 1990s is the most dominant ESBL among Enterobacteriaceae and is recognized as a rapidly growing family of ESBLs. The CTX-M enzyme is the predominant type of ESBL found in many regions of the world, including Asia, South America, Europe and Africa. The CTX-M enzyme forms a rapidly growing family of over 69 enzymes currently [7]. This study has been undertaken to identify the occurrence of ESBL among Escherichia coli and Klebsiella pneumoniae from cases of VAP and detect the genes responsible for ESBL production by conventional PCR method.
Material and Methods
A prospective study was done over a period of 5 years from 2006 to 2010 in a urban city, Tumkur, Karnataka, India, which included patients who were clinically diagnosed as VAP [8]. Ethical clearance was obtained before collection of samples. Endotracheal Aspirates (EA) [9] were collected from patients fulfilling the clinical criteria for VAP. EA were cultured by quantitative method [10] and presence of >105cfu/ml was considered. The bacterial isolates were identified to species level by standard biochemical tests [11].
Antibiotic Susceptibility Test
Antibiotic susceptibility testing was done by Kirby Bauer Disk Diffusion method on Mueller Hinton Agar as per Clinical and Laboratory Standard Institute guidelines (CLSI) [12] to determine the resistance patterns of the isolates. Escherichia coli ATCC 25922 was used as the control. Samples showing an inhibition zone size of ≤ 22 mm with ceftazidime and ≤ 27 mm with cefotaxime were considered as potential ESBL producer and were further investigated for confirmation.
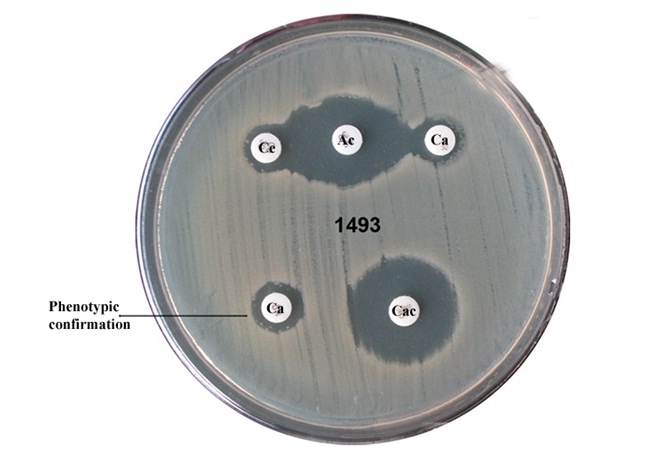
Detection of ESBL by Disk Approximation method [
13]
Isolates that showed intermediate/resistance to 3rd generation cephalosporins were screened to detect ESBL production. A modified double disk synergy (Disk Approximation Test) was carried out on resistant isolates. Amoxicillin/clavulanic acid (20μg/10μg) disk was placed in the centre of the MHA plate on which a lawn culture of the test organism (turbidity matched to 0.5 McFarland turbidity) had been made, ceftazidime (Ce) (30μg) and cefotaxime (Ca) (30μg) were placed on either side at a distance of 15 mm centre to centre from the amoxicillin/clavulanic acid (Ac) disc. Plates were incubated at 35oC for 18-24 hrs and the pattern of zone of inhibition was noted. Isolates that exhibited a distinct potentiation towards amoxicillin + clavulanic acid disc were considered potential ESBL producer. Escherichia coli ATCC 25922 and Klebsiella pneumoniae ATCC 700603 were negative and positive controls respectively.
Combination Disc Diffusion Method [
12]
A cefotaxime 30μg and a cefotaxime + clavulanic acid (30μg+10μg) discs (Hi-media, Mumbai) were placed at a distance of 25 mm on a Mueller-Hinton Agar plate incubated with a bacterial suspension of 0.5 McFarland turbidity standards and incubated overnight at 37oC. A ≥ 5mm increase in the diameter of inhibition zone for the combination disc versus ceftazidime disc confirmed ESBL production.
ESBL producers among the isolates were subjected to PCR for gene detection. The Boiling method was used to extract DNA from Bacterial samples. TEM, SHV and CTX-M beta-lactamase genes were detected by PCR. PCR were carried out using thermal cycler (Eppendorf Mastercycler Gradient). The PCR mix was prepared in a volume of 20 μl containing 1 μl DNA template, 2 ml of 10 X PCR buffer (Sigma, Aldrich), 1.5 mM MgCl2 (Sigma, Aldrich), 0.6 μl 2 mM dNTPs (ferments), 1 U taq polymerase (Sigma, Aldrich), 1μl of 10 pmol of each three primers (Eurofins, Bangalore), volume made up to 20 μl with distilled water. The primer sequences and cycling conditions used and product size for three different PCR are shown in [Table/Fig-1 & 2]. The PCR products were separated by gel electrophoresis on 1% agarose gel. Escherichia coli Inhouse strains positive for bla TEM, bla SHV and bla CTX-M were used as positive controls and Escherichia coli ATCC 25922 were used as negative control. For all the PCR initial denaturation and final extension were 94oC for 4 min and 72oC for 8 min respectively.
Primers used for amplification of bla TEM, bla SHV, bla CTX-M genes
| Sl. No. | Gene Detected | Primer |
|---|
| 1 | bla TEM1[15] | F 5’ ATAAAATTCTTGAAGACGAAA 3’ R 5’ GACAGTTACCAATGCTTAATCA 3’ |
| 2 | bla SHV[15],[16] | F 5’ GGGTAATTCTTATTTGTCGC 3’ R 5’ TTAGCGTTGCCAGTGCTC 3’ |
| 3 | bla CTX-M[17] | F 5’ TTTGCGATGCAGTACCAGTAA 3’ R5’CGTATATCGTTGGTGGTGCCATA 3’ |
PCR Conditions for amplification of bla TEM, bla SHV, bla CTX-M genes bp: base pair
| Gene Detected | Denaturation Time/temp | Annealing Time/temp | Extension Time/temp | No. of cycles | Product |
|---|
| bla TEM | 94oC-1min | 58oC -1 ½ min | 72oC -1min | 30 | 1100bp |
| bla SHV | 94oC-1min | 58oC -1 ½ min | 72oC -1min | 30 | 930bp |
| bla CTX-M | 94oC-1min | 58oC -1 ½ min | 72oC -1min | 30 | 544bp |
Results
In this prospective study, among the isolates from 428 patients who developed VAP, 144 isolates belonged to the Enterobacteriaceae family (Klebsiella pneumoniae 87 and Escherichia coli 57) 428 patients developed VAP. In this initial screening by standard disc diffusion method 86 isolates were identified as ESBL producers (59.72%). A total of 66 isolates (28 of Klebsiella pneumoniae and 38 of Escherichia coli) were confirmed as ESBL producer by disc approximation method and 63 isolates by double disc combination method [Table/Fig-3]. [Table/Fig-4] shows the distribution of ESBL.
Disk Approximation method
| Sl. No | Isolates | No. isolated | ESBL |
|---|
| | | KB disc Diffusion Method | Disk Approximation Method | Combination Disc Method |
| 1. | Klebsiella pneumoniae | 87 | 37 (42.5%) | 26 (29.8%) | 28 (32.1%) |
| 2. | Escherichia coli | 57 | 49 (85.9%) | 37 (64.9%) | 38 (66.6%) |
| Total | 144 | 86 (59.7%) | 63 (43.7%) | 66 (45.8%) |
Among 38 Escherichia coli, 30 (78.95%) showed the presence of at least one ESBL genes. The distribution of more than one gene were seen in four isolates having a combination of genes bla TEM + bla CTX-M in two strains and bla SHV + bla CTX-M in two strains [Table/Fig-5, 6 & 7]. Among 28 Klebsiella pneumoniae phenotypically confirmed as ESBL producer only single gene was detected in 24 (85.71%) isolates. [Table/Fig-8] shows the distribution of ESBL genes.
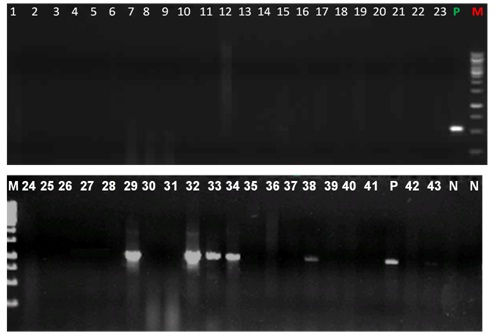
PCR for bla TEM GENE M - Marker 250bp-3000bp No. 1-43 isolates lanes 4,5,8,10,11,18,19,25,26,28,29,33 Klebsiella pneumoniae Lanes 1-3,12-17,21-24,30-32,34-43 Escherichia coli.
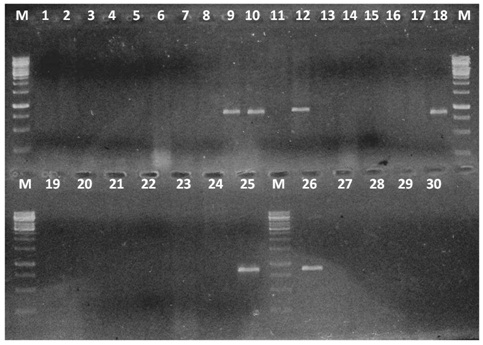
PCR for bla SHV GENE M - Marker 250bp-3000bp No. 1-43 isolates lanes 4,5,8,10,11,18,19,25,26,28,29,33 Klebsiella pneumoniae Lanes 1-3,12-17,21-24,30 Escherichia coli
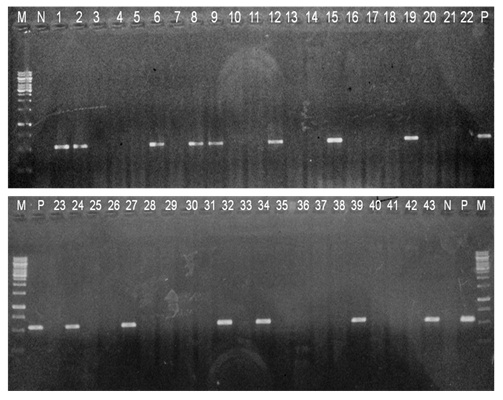
PCR for bla CTX-M GENE M - Marker 250bp-3000bp No. 1-43 isolates lanes 4,5,8,10,11,18,19,25,26,28,29,33 Klebsiella pneumoniae Lanes 1-3,12-17,21-24,30-32,34-43, Escherichia coli
Distribution of ESBL genes
| Sl. No. | Isolate | bla TEM | bla SHV | bla CTX-M | bla TEM+ bla CTX-M | bla SHV+ bla CTX-M |
|---|
| 1. | Klebsiella pneumoniae | 02 | 10 | 12 | 00 | 00 |
| 2. | Escherichia coli | 04 | 02 | 20 | 02 | 02 |
Discussion
Increasing resistance to third generation cephalosporins has become a cause of concern especially among Enterobacteriaceae that cause nosocomial infections including VAP. The prevalence of ESBL among members of Enterobacteriaceae constitutes a serious threat to current β lactam therapy leading to treatment failure and consequent escalation of costs of hospital stay. Among the wide array of antibiotics, β lactam are the most widely used for over 50% of all the systemic infections [4].
In our study 46.15% ESBL producers belonged to Enterobacteriaceae family. There have been reports of ESBL’s from major hospitals in India and some of them have recorded the incidence to be as high as 60-68% [18, 19]. The high incidence of ESBL is a cause of concern to regulators of hospital antibiotic policy. Over reliance on third generation cephalosporins to treat gram negative bacterial infections is one of the prime causes for increased resistance to this class of antibiotics.
The difference observed in detection of ESBL positive isolates by two different methods may be justified by the lower sensitivity of phenotypic method and the influence of environmental factors on the incidence of resistance [20]. The lack of constant sensitivity of different phenotypic methods has been emphasized by some studies [21]. In contrast, the genotypic method using specific PCR amplification of resistance genes seems to have 100% specificity and sensitivity. The cost of molecular method is particularly reduced for the bacteria belonging to enterobactericeae family as their DNA is easily extractable by boiling method, a quick and cost effective DNA extraction method [20].
The ESBL producers usually carry multiple resistant plasmid. In our study 4 isolates of Escherichia coli bla TEM + bla CTX-M and bla SHV+ bla CTX-M. Similar results have been reported by Yazdi M et al., and Yuan et al., [20, 22]. Most ESBLs are derived from plasmid mediated penicillinases belonging to the TEM and SHV families. Recently the CTX-M group with a typical ESBL resistant phenotype which does not originate from TEM and SHV families has been described. The CTX-M group is a new family of plasmid mediated ESBL they preferably hydrolyse cefotaxime have been recognized by Xiong et al., [23].
In the present study bla CTX-M was the common gene in 48.5% strains followed by 22.22% bla SHV and 14.81% bla TEM. Similar results have been obtained in many studies in India and other countries [7]. In a study conducted from 10 Indian sites in six Indian cities by Welsh et al., [24] bla CTX-M was the most common genotype isolated. Similar results have been documented in Europe, Latin America and other countries [25]. The CTX-M genotype, originating from chromosomally encoded enzymes of the Kluyvera spp, has risen in predominance especially in Escherichia coli and Klebsiella pneumoniae they have greater ability to spread and cause outbreaks [7].
The presence of more than one gene type in some of the isolates like bla TEM+ bla CTX-M means that the ESBL producing strains may be related to complex antimicrobial resistance. bla TEM producing TEM-1 is a broad spectrum β-lactamase that is always combined with CTX-M in the same plasmid [26]. It is important to continuously monitor resistance trends and enhancing the infection control of these pathogens in health care units.
Conclusion
Our findings suggest that the presence of bla CTX-M as the predominant genotype in Klebsiella pneumoniae and Escherichia coli isolates even in small town as ours. This study documents the presence of CTX-M type β lactamase even in hospitals of a small urban town in Karnataka. Incorrect identification of antibiotic resistance may lead to inappropriate antibiotic prescription, which in turn may direct bacteria to produce new resistance genes by selective pressure. The high incidence of ESBL is a cause of concern to regulators of hospital antibiotic policy. The genotypic method using specific PCR amplification of resistance genes seems to have 100% specificity and sensitivity. Therefore, the genotypic method is suggested to be used as the method of choice for detection of ESBL producing strains.
[1]. Chastre J, Fagon JY, Ventilator associated pneumoniaAm J Respir Crit Care Med 2002 165:867-903. [Google Scholar]
[2]. Safdar N, Dezfulian C, Collard HR, Saint S, Clinical and economic consequences of ventilator associated pneumonia: A systematic reviewCrit care Med 2005 33:2184-93. [Google Scholar]
[3]. Rello J, Ollendrof DA, Oster G, Vera-Llonch M, Redman R, Epidemology and outcomes of ventilator associated pneumonia in a large US databaseChest 2002 122:2115-21. [Google Scholar]
[4]. Kumar RS, Lakshmi V, Rajagopal R, Occurence of extended spectrum Beta-lactamase among Enterobacteriaceae spp. isolated at a tertiary care instituteIndian J Med Microbiol 2006 24(3):208-11. [Google Scholar]
[5]. Bazzaz BS, Naderinasab M, Mohamadpoor AH, Farshadzadeh Z, Ahmadi S, Yousefi F, Escherichia coli and Klebsiella pneumoniae among clinical isolates from a general hospital in IranActa Microbiol Immunol Hung 2009 56(1):89-99. [Google Scholar]
[6]. Asgar AH, Prevalence and Potential Types of Extended Spectrum β-Lactamases among Escherichia coli and Klebsiella species in Makkah HospitalsOxford Research Forum Journal 2007 2(4):47-55. [Google Scholar]
[7]. Al-Algamy MHM, Shibl AM, Tawfik AF, Prevalence and molecular characterization of extended-spectrum β-lactamase-producing Klebsiella pneumoniae in Riyadh, Saudi ArabiaAnn Saudi Med 2009 29(4):253-07. [Google Scholar]
[8]. Dwivedi M, Mishra A, Azim A, Singh RK, Baroni AK, Prasad KN, Ventilator-associated pneumonia caused by carbaoenem-resistant Enterobacteriaceae carrying multiple metallo-beta lactamase genesIndian J Pathol Microbiol 2009 52(3):339-42. [Google Scholar]
[9]. Rajasekhar T, Anuradha K, Suhasini T, Lakshmi V, The role of quantitative cultures of non-bronchoscopic samples in ventilator-associated pneumoniaIndian J Med Microbiol 2006 24(2):107-13. [Google Scholar]
[10]. Baselski VS, El-Torky M, Coalson JJ, Grifin JP, The Standardization of Criteria for processing and Interpretation of Laboratory specimens in patients with suspected ventilator associatedChest 1992 102(5):571-79. [Google Scholar]
[11]. Collee JG, Miles RS, Tests for Identification of Bacteria. In: Collee JG, Duduid JP, Fraser AG, Matmion, editorsMackie and McCartney, Practical Medical Microbiology 1996 14th edNew YorkLongman Singapore publishing:141-60. [Google Scholar]
[12]. Clinical Laboratory Standard InstitutePerformance Standards for antimicrobial disk susceptibility tests approved standard 2012 9th edWayne PACLSILSI document M2-49 [Google Scholar]
[13]. Jarlier V, Nicholas NH, Fourier G, Phillippon Extended broad spectrum β lactamase conferring transferable resistance to newer β lactam agents in Enterobacteriaceae, hospital prevalence and susceptibility patternsRev Infect Dis 1988 10(4):867-78. [Google Scholar]
[14]. Sambrook Russell In Molecular Cloning: A Laboratory Manual 2007 vol 13 edNew YorkCSHI Press:1.3.2-1.3.4. [Google Scholar]
[15]. Siu LK, Lu PL, Hsueh PR, Lin FM, Chang SC, Luh KT, Bacteraemia due to extended spectrum β lactamase - Producing Escherichia coli and Klebsiella pneumoniae in Pediatric Onchology ward: Clinical Feature and Identification of Different Plasmids carrying both SHV5 and TEM1 GenesJ Clin Microbiol 1999 37(12):4020-07. [Google Scholar]
[16]. Oliveira CF, Salla A, Lara VM, Rieger A, Horta JA, Alves SH, Prevalence of extended spectrum β lactamase – producing micro-organism in nosocomial patients and molecular characterization of the SHV type isolatesBraz J Microbiol 2010 41(2):1517 [Google Scholar]
[17]. Edelstein M, Pimkin M, Palagin I, Fdelsteini Stratchounski Prevalance and Molecular Epidemiology of CTX-M Extended Spectrum β lactamase producing Escherichia coli and Klebsiella pneumoniae in Russian HospitalsAntimicrob Agents Chemother 2003 47(12):3724-32. [Google Scholar]
[18]. Mathur P, Kapil A, Das B, Dhawan B, Prevalence of extended spectrum beta lactamase producing bacteria in a tertiary care hospitalIndian J Med Res 2002 115:153-7. [Google Scholar]
[19]. Jones RN, Rhomberg PR, Varman DJ, Mathai D, A comparasion of antimicrobial activity of meropenem and selected broad-spectrum antimicrobials tested against multidrug resistant Gram negative bacilli including bacteriaemia Salmonella spp: Initial studies for MYSTIC programme in IndiaInt J Antimocrob Agents 2002 20:426-31. [Google Scholar]
[20]. Yazdi M, Nazemi A, Mirinargasi M, Jafarpour M, Sharifi SH, Genotyping versus Phenotyping methods to detect Extended-Spectrum Beta-Lactamase (ESBLs) in uropathogenic Escherichia coliAnnals of Biological Research 2012 3(5):2454-58. [Google Scholar]
[21]. Ravi SG, Namratha WN, Krishna BVS, Chandrashekar MR, Comparison of Disc Diffusion Methods for the Detection of Extended-Spectrum Beta Lactamase-Producing EnterobacteriaceaeJ Lab Physicians 2011 3(1):33-06. [Google Scholar]
[22]. Yuan X, Jianfei C, Yuping H, Resistance of Strains Producing Extended-Spectrum β-lactamase and Genotype Distribution among Escherichia coli in ChinaPakistan J Zool 2012 44(2):457-21. [Google Scholar]
[23]. Xiong Z, Zhu D, Wang F, Zhang Y, Okamoto R, Inoue M, Klebsiella pneumoniae producing three kinds of class A β lactamase encoded by one single plasmid isolated feom a patient in Huashan Hospital, Shanghai, ChinaInt J Antimicrobial Agents 2002 23:262-07. [Google Scholar]
[24]. Welsh TR, Toleman MA, Jones RN, Comment on: Occurrence, prevalence and genetic environment of CTX-M β lactamase in Enterobacteriaceae from Indian hospitalsJ Antimicrob Chemother 2006 58(6):1260-03. [Google Scholar]
[25]. Dhillon RHP, Clark J, ESBL’s: A clear and Present DangersCrit Care Res Pract 2012 [Google Scholar]
[26]. Ye QH, Lau Y, Liang B, Tian SF, Antimicrobial resistance, genotypic charateristization and pulse field gel electrophoresis typing of extended spectrum β lactamase-producing clinical Escherichia coli strains in Macao, ChinaChin Med J (Engl) 2011 124(17):2701-07. [Google Scholar]