Sepsis is the commonest cause of neonatal mortality and is probably responsible for 30-50% of total neonatal deaths each year in developing countries [1]. It is estimated that 20% of all neonates develop sepsis and approximately 1% die of sepsis related causes [2]. National neonatal perinatal database from India has reported an incidence of neonatal sepsis of 0.1% to 4.5% from 18 hospitals across India and the incidence of blood culture proven sepsis was reported as 8.5 per 1,000 live births [3,4].
Neonatal sepsis can be divided into two main classes depending on the onset of symptoms namely Early-onset sepsis, which usually presents within the first 72 hours of birth and late-onset sepsis, which usually presents 72 hours after birth [2].
The pattern of organisms causing neonatal sepsis has been constantly changing. Compounding the problem is the frequent emergence of resistant bacteria in the nurseries [5].
During the past decade, ESBL strains have frequently been implicated in neonatal intensive care units at tertiary care hospitals. Recent study involving four different centres in India showed that Klebsiella species and Escherichia coli are the most common GNB causing neonatal sepsis in India and one-third are ESBL producers in both community and hospital settings and is associated with very high mortality rate (33%) in these patients [1,6]. MRSA has become one of the most common pathogens in NICU with 33% prevalence as per a recent Indian study [7].
The emergence of multidrug resistant bacteria points to the importance of antibiotic policies for the neonatal intensive care unit.
Therefore, this study was designed to study the bacteriological profile in neonatal sepsis and to study their antimicrobial susceptibility pattern including detection of methicillin resistance in Staphylococci and ESBL, AmpC-beta-lactamases and metallo-beta-lactamase production in Gram negative bacteria.
Materials and Methods
This study was conducted for a period of one and a half years from January 2010 to June 2011 in a tertiary care hospital in Chennai, India, after obtaining due approval from the institutional ethics committee. A total of 182 blood samples were collected during the study.
Inclusion criteria: Neonates clinically suspected of having sepsis, temperature >99°F or <95°F, respiratory rate>60 per minute, abnormal cry, refusal of feed, drowsy or unconscious, septic focus on skin or umbilicus, diarrhea and seizures.
Exclusion criteria: Neonates already on antibiotics and with diagnosis of intra-uterine infection and congenital anomalies were excluded from the study.
Specimen Processing
Blood samples were collected from fresh veni-puncture site after preparing the area with povidone–iodine and 70% alcohol and were subjected to aerobic bacterial culture by conventional method using brain heart infusion broth with sodium polyanetholsulphonate (Himedia Labs, India). Another 1 ml of collected blood was allowed to clot in a dry testtube, serum separated and used for CRP estimation [Table/Fig-1]. Bacteria isolated by standard microbiological procedures were subjected to antimicrobial susceptibility testing by Kirby Bauer’s disc diffusion method. Suspected multidrug resistant strains were confirmed by phenotypic and genotypic methods. Relevant ATCC strains were used as controls wherever necessary.
Correlation of CRP positivity with culture positivity
| CRP Positive | CRP Negative |
|---|
| Culture positive | 94(94.9%) | 16 (19.3%) |
| Culture negative | 5(5%) | 67(80.7%) |
Phenotypic detection of MRSA [8]: [Table/Fig-2]
| Organism | MRSA using Cefoxitin disc (30 μg) | MRSA using Oxacilin disc (1μg) |
|---|
| Staphylococcus aureus (n=30) | 17(56.6%) | 15(50%) |
Detection of methicillin resistance among Staphylococcus aureus isolates was done using 1 μg oxacillin disc on Mueller Hinton agar supplemented with an additional 5% NaCl and Cefoxitin disc (30 ug) diffusion test and results interpreted according to CLSI guidelines.
Phenotypic detection of ESBL production [5,9] [Table/Fig-3]
ESBL and AmpC detection in Enterobacteriaceae n=49
| Isolates | ESBL | AmpC | Co- existence of ESBL and AmpC |
|---|
| Escherichia coli (n=14) | 8 | - | - |
| Klebsiella pneumoniae (n=31) | 22 | 3 | 3 |
| Klebsiella oxytoca (n=4) | 3 | - | - |
| Total % (n=49) | 33 (67.3%) | 3 (6.1%) | 3 (6.1%) |
Gram negative isolates that were resistant to at least 2-3rd generation cephalosporin namely cefotaxime (30μg) and ceftazidime (30μg) were considered to be probable ESBL producers and further confirmation was done by the Phenotypic Confirmatory Test for ESBL Production- Combined Disc Test [Table/Fig-4].
Combined disc test for ESBL, AmpC disc test
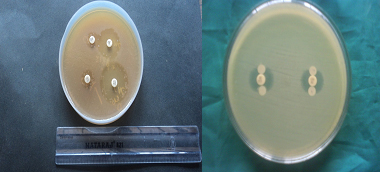
MIC was determined by agar dilution method [10] for all probable ESBL producers. Isolates were tested for various concentrations of cephalosporin alone (0.5μg – 2048μg/ml of agar) and in combination with 4μg /ml of clavulanic acid from 0.5μg to 2048μg / ml of agar and the MIC determined. Three times doubling dilution reduction in the MIC of 3rd generation cephalosporins in the presence of clavulanic acid was considered as production of ESBL [11].
Molecular Detection of ESBL production: (Conventional Polymerase chain reaction) [Table/Fig-5,6].
PCR for detection of ESBL gene. Molecular Marker-100bp, Lane 1 to 13 –Sample, Gel % -1.2 %

Primers and thermal cycling conditions that were used for amplification of TEM, SHV and CTX-M genes [12]
| Target gene | Primers used | Thermal cycling conditions |
|---|
| blaTEM | GTATCCGCTCATGGAGACAATAACCCTG CCAATGCTTAATCAGTGGAGGCACC | 95°C 15 min → 30x[95°C 1min,58 °C 1 min, 72 °C 1 min] → 72 °C 10 min → 4 °C ∞ |
| blaSHV | CGCCTGTGTATTATCTCCCTGTTAGCC TTGCCAGTGCTCGATAGAC | 95°C 15 min → 30x[95°C 1min,58 °C 1 min, 72 °C 1 min] → 72 °C 10 min → 4 °C ∞ |
| blaCTX-M | CGCTTTGCGATGCGAG ACCGCGATATCGTTG | 95°C 15 min → 30x[95°C 30 sec,50 °C 40 sec, |
Detection of AmpC -AmpC disc test [13]
The test is based on use of Tris-EDTA to permeabilize a bacterial cell and release β-lactamases into the external environment. AmpC discs were prepared in-house by applying 20 μl of a 1:1mixture of saline and 100 x Tris-EDTA to sterile filter paper discs, allowing the discs to dry, and storing them at 2 to 8°C. The surface of a Mueller-Hinton agar plate was inoculated with a lawn of cefoxitin susceptible Escherichia coli ATCC 25922 according to the standard disc diffusion method. Immediately prior to use, Amp C discs were rehydrated with 20 μl of saline and several colonies of each test organism were applied to a disc. A 30- μg cefoxitin disc was placed on the inoculated surface of the Mueller-Hinton agar. The inoculated Amp C disc was then placed almost touching the antibiotic disc with the inoculated disc face in contact with the agar surface. The plate was then inverted and incubated overnight at 35°C in ambient air. After incubation, plates were examined for either an indentation or a flattening of the zone of inhibition, indicating enzymatic inactivation of cefoxitin (positive result), or the absence of a distortion, indicating no significant inactivation of cefoxitin (negative result).
Detection of MBL
MBL Demonstration [14]
MBL production was demonstrated by two methods namely Combined disc test with EDTA and Ceftazidime (30μg) - EDTA Double disc synergy test.
C - Reactive protein Estimation
CRP estimation was done using CRP-latex agglutination slide test kit.
Test Statistics used: For continuous variables- mean, standard deviation, p-value using ‘t’ test. For discretenominal variables – Chi - square.
Results
There were 182 clinically suspected cases of neonatal sepsis. A culture positivity rate of 110 (60.4%) was observed. Culture positivity in pre-terms was 14 (53.8%). There were 141 (77.5%) cases of early onset neonatal sepsis out of which 86 (60.99%) were culture positive. There were 41 (22.5%) Late onset sepsis cases out of which 24 (58.5%) were culture positive. Early onset sepsis cases showed presence of majority of Gram negative isolates (63.9% of total Early onset sepsis cases). In cases of late onset sepsis, it was predominantly due to Gram positive organisms- 19 (79.1%). As per [Table/Fig-1], CRP was positive in 99 (54.3%) of the total 182 cases. Out of the total 99 CRP positive cases, 94 (94.9%) were culture positive with a significant p-value (<0.0000001) and odds ratio 78.72. Most common pathogen identified was Klebsiella pneumoniae- 31(28%) followed by Staphylococcus aureus-30 (27%), Escherichia coli-14 (12.7%), Coagulase negative Staphylococci-12 (10.9%), Pseudomonas aeruginosa-11 (10%), Enterococcus faecalis-8 (7.2%) and Klebsiella oxytoca-4 (3.63%), 22 (71%) of Klebsiella pneumoniae isolates were sensitive to Amikacin and 21 (67.7%) of them were sensitive to Gentamicin and Ofloxacin. For Imipenem, 100% sensitivity was noted.
Among the Escherichia coli isolates, higher sensitivity was noted for Imipenem-14 (100%). Eleven (78.6%) of them were sensitive to Gentamicin, Amikacin and Ofloxacin 9 (81%) of Pseudomonas aeruginosa isolates were sensitive to Ciprofloxacin and Ofloxacin. Eight (72.7%) of them were sensitive to Amikacin. Metallobetalactamase producers among Pseudomonas aeruginosa was 3 (27.2%).
Discussion
Neonatal sepsis is a major contributor to neonatal mortality and has to be addressed seriously. World Health Organization has estimated that 1.6 million deaths occur globally every year due to neonatal infections and 40% of them occur in developing countries [4]. In the present study, 182 neonates suspected of having sepsis were investigated. National Neonatal Perinatal database (NNPD) 2003 [3] classifies sepsis as early onset sepsis (presenting within 72 hours of birth) and late onset sepsis (presenting 72 hours after birth). Age of the babies ranged from 1 day to 21 days. Most of the babies had clinical presentation of breathlessness, poor cry and poor activity. Other manifestations included poor feeding, convulsions, fever, sclerema, jaundice, tachypnoea, grunt, lethargy, irritability, diarrhea and abdominal distension. In our study, the most common pathogen identified was Klebsiella pneumoniae 31(28%). This is similar to study by Zakariya et al., [4], and Chandel et al., [6]. NNPD [3] data 2002-2003 shows that Klebsiella pneumoniae is the commonest pathogen isolated in neonatal sepsis both in babies born within the institution (intramural) and in babies referred from community and other hospitals (extramural).
In our study, as per [Table/Fig-1], CRP was positive in 99 (54.3%) of the total 182 cases, out of which 94 (94.9%) were culture positive. Many studies have stressed the importance of CRP in neonatal sepsis. It is helpful in following the course and prognosis of the disease. The early evaluations of CRP provide indications of the response to the treatment. A good response to antibiotics is indicated by a rapid return to normal of CRP [15].
In our study,as per [Table/Fig-2], there were a total of 30 (27%) Staphylococcus aureus isolated, out of which 17 (56%) were methicillin resistant. All the MRSA strains were sensitive to vancomycin. Enterococcus faecalis and Coagulase negative Staphylococci were also 100% sensitive to Vancomycin. This is similar to Karthikeyan et al., [16] who reported that 66% of Staphylococcus aureus isolated from cases of neonatal sepsis were methicillin resistant. Isolation of MRSA was 31.25% as per Gandhi et al., [17] and it was 33% according to Patel et al., [7]. So screening for MRSA in every Staphylococcus aureus isolated will be of immense value for providing efficient patient care.
The widespread use of broad spectrum β-lactam antibiotics has led to a marked increase in the incidence of ESBL in Gram negative organisms [18]. As per [Table/Fig-3], ESBL were detected in 33 (67.3%) of Enterobacteriaceae. This is similar to study from North India [19] in which 61.5% of Gram negative isolates were ESBL producers.
ESBL isolates were 100% sensitive to Imipenem. Greater resistance to beta lactam and non-beta-lactam antibiotics was evident in ESBL producers than in those, which were ESBL negative. This is similar to Bhattacharjee et al., [1].
In our study, ESBL was detected in 70.9% of Klebsiella pneumoniae isolates and 57.14% of Escherichia coli isolates. This is closer to study by Bhattacharjee et al., [1] in which ESBL was observed in 62.7% Klebsiella pneumonia and 46.5% of Escherichia coli and Jain et al., [5] in which ESBL was in 86.6% of Klebsiella pneumoniae and 63.6% of Escherichia coli. As per Gandhi et al., [17], ESBL production was seen in 52.9% of Escherichia coli and 50% of Klebsiella pneumoniae isolates. Conventional PCR was done and out of the 49 Enterobacteriaceae, TEM gene was detected in 11 (22.4%). TEM gene occurred in 3 (37.5%) of Escherichia coli, 7 (31.8%) of Klebsiella pneumoniae and 1 (33.3%) of Klebsiella oxytoca.
As per [Table/Fig-3], AmpC beta lactamases was detected in 3 (6.1%) of Enterobacteriaceae, and there was co-existence of ESBL and Amp-C beta lactamases in those isolates. This is similar to observation made by Singhal et al., [20], which showed Amp-C beta lactamases in 8%isolates.
Isolates with ESBL and AmpC beta lactamases yielded higher MICs than other isolates. Chatterjee et al.,[21] has reported that there is high prevalence of co-expression of ESBLs, Amp-C beta lactamases and MBL in Gram negative bacilli. The presence of ESBLs and Amp-C beta lactamases in a single isolate reduces the effectiveness of the β-lactam-β-lactamase inhibitor combinations, while MBLs and Amp-C beta lactamases confer resistance to carbapenems. Often these enzymes are co-expressed in the same isolate [21]. MBL producers among Pseudomonas aeruginosa was 3 (27.2%). The high percentage of ESBL producing isolates may be due to the selective pressure imposed by extensive use of antimicrobials in intensive care units. The difference in percentage of isolation of ESBL compared to few other studies could be due to regional variations. Distinct regional variations have been detected in the incidence of ESBL producing isolates [5]. Recent study demonstrated that restricted use of cephalosporins significantly reduced the incidence of ESBL producing, cefotaxime resistant and ciprofloxacin resistant gram negatives [22].
Simple hygienic measures, such as hand washing practices, the use of sterile equipment, patient cohorting (i.e. grouping patients with similar infections in the same location) and screening of attending staff and mothers for MRSA and ESBL can help prevent the further spread of these resistant strains. This study stresses that antimicrobial resistance is a global problem and emphasizes the need for surveillance and promotion of correct and restrictive antibiotic policies including specific antibiotic therapy after studying sensitivity pattern. This will halt the further spread of MRSA and ESBL and improve the prognosis in cases of neonatal sepsis.
Conclusion
Neonatal sepsis is an important cause of neonatal mortality and it depends upon the age of onset of sepsis, upon the etiologic agent and their resistant pattern. Implementation of infection control measures, restricting the use of broad spectrum antibiotics,rotation of antibiotics and rationalizing the use of antibiotics can decrease antibiotic resistance. Early diagnosis and specific treatment can reduce neonatal mortality and morbidity.